SUPPLEMENTARY WEB-PAGE
An Empirical Bayes Adjustment to Increase the Sensitivity of Detecting
Differentially Expressed Genes in Microarray
Experiments
Susmita Datta1, Glen A. Satten2,
Dale J. Benos3, Jiazeng Xia3,
Martin J. Heslin4, Somnath Datta* 5
1 Department of Mathematics and Statistics, Georgia State University,
Atlanta, GA 30303.
2 Centers for Disease Control and Prevention, Atlanta, GA
30333.
3 Department of Physiology and Biophysics, University of Alabama at Birmingham, Birmingham, AL 35294.
4 Department of Surgery, University of Alabama at Birmingham,
Birmingham, AL 35294.
5 Department of Statistics, University of Georgia,
Athens, GA 30602.
* Corresponding author:
somnath.datta@louisville.edu
Bioinformatics, 20, 235-242 (2004)
Motivation: Detection of differentially expressed genes is one of the major
goals of microarray experiments. Pairwise
comparison for each gene is not appropriate without controlling the overall (experimentwise) type 1 error rate. Dudoit et al. have advocated use of permutation-based
step-down P-value adjustments to correct the observed significance levels for
the individual (i.e., for each gene) two sample t-tests.
Results: In this paper, we consider an ANOVA formulation of the gene expression
levels corresponding to multiple tissue types. We provide resampling-based
step-down adjustments to correct the observed significance levels for the
individual ANOVA t-tests for each gene and for each pair of tissue type
comparisons. More importantly, we introduce a novel empirical Bayes adjustment to the t-test statistics that can be
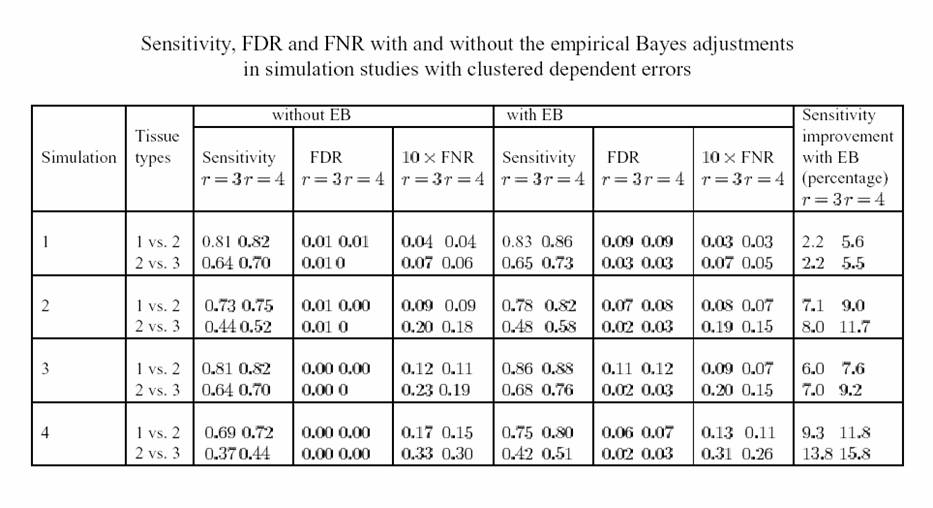
incorporated into the step-down procedure. Using simulated data, we show
that the empirical Bayes adjustment improved the
sensitivity of detecting differentially expressed genes up to 16%, while
maintaining a high level of specificity. This adjustment also reduces the FNR
(false non-discovery rate) to some degree at the cost of a modest increase in
the FDR (false discovery rate). We illustrate our approach using a human colon
cancer data set consisting of oligonucleotide arrays
of normal, adenoma and carcinoma cells. The number of genes with differential
expression level declared statistically significant was about fifty when
comparing normal to adenoma cells and about five when comparing adenoma to
carcinoma cells. This list includes genes previously known to be associated
with colon cancer as well as some novel genes.
##################################################################################
Data set (colon cancer) download
For further details on the data set
contact Dr. Dale Benos (benos@physiology.uab.edu)
##################################################################################
R-code used to calculate
the P-values of the empirical Bayes adjusted t-tests
for the colon cancer data download
This is being distributed without warrantee; use at your own risk!
##################################################################################
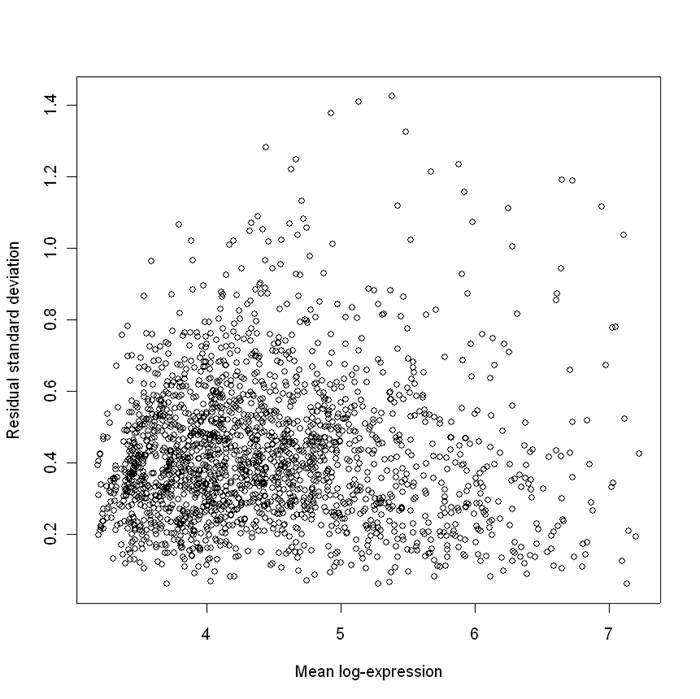
Mean (log) expression level versus standard deviation of the residuals for the colon cancer data:
For easy visualization, values for the top 2000 (most differentially expressed) genes are plotted.
##################################################################################
##################################################################################